

We are dedicated to providing outstanding customer service and being reachable at all times.
Viral Genome De Novo Sequencing
CD Genomics is a leading custom service provider in sequencing technologies, particularly using next-generation sequencing (NGS) and long-read sequencing technologies. We offer viral genome de novo sequencing service which is part of our integrated portfolio of microbial whole-genome de novo sequencing services. Based on our advanced platform and highly skillful scientists, we can assist researchers and professionals investigate unexplored taxa or studying viruses with high mutation and/or recombination rates.
De novo assembly is one of the main bioinformatic strategies for genome assembly, referring to the generation of contigs without reference genomes. Sequencing technologies and de novo assembly are essential to generating complete viral genomes, providing important information on virus transmission, characterizing emerging viruses, and exploring genomic regions with important functions for evading the host immune system or antiviral drugs.
Viral Genome De Novo Sequencing Service in CD Genomics
Our service platform combines short-read (Illumina) and long-read sequencing (ONT and PacBio SMRT) to quickly and efficiently generate high-resolution sequence data and improve the assembly of a variety of viral genomes. Short-read sequencing can provide data with high throughput and relatively low error rates. While long-read sequencing can offer more accurate, longer, and more affordable long reads. More importantly, RNA viral genomes can be directly sequenced avoiding bias and requiring no assembly. Therefore, this makes it possible to analyze epigenetic modifications alongside nucleotide sequence.
Our Service Support
- Combination of short and long-read sequencing without reference sequences.
- Complete characterization of viral genomes.
- Generate complete de novo assemblies of large viral genomes.
Sample Requirements
- Sample Type: Genomic DNA/cDNA, OD260/280=1.8~2.0, no degradation and no contamination.
- Viral DNA amount:
-Illumina platform: ≥ 1 μg.
-PacBio/ONT platform: ≥ 5 μg.
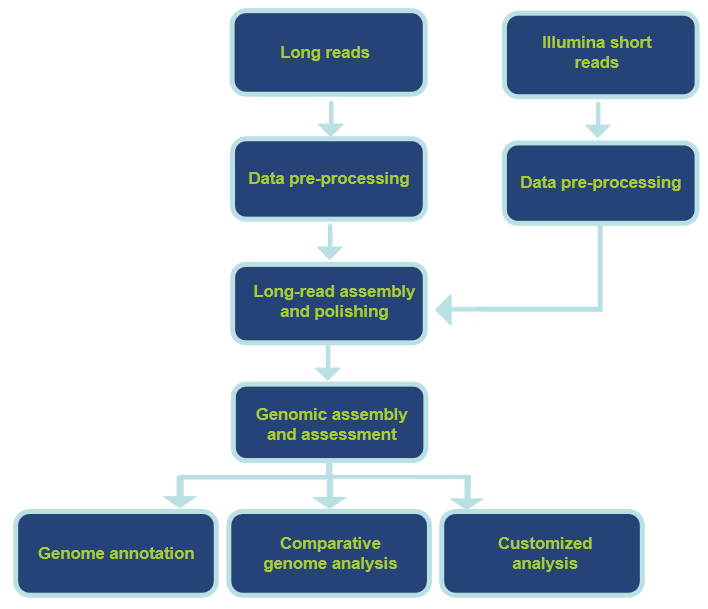
Analysis Workflow
Analysis Content
| Genome Survey | Genome Assembly | Genome Annotation |
| -Genome size assessment -Genome complexity assessment -Genome pre-assembly |
-De novo genomic assembly -Assembly outcome evaluation |
-Repetitive sequence analysis -Gene structural annotation -Gene functional annotation -Non-coding RNA annotation |
Service Highlights
- Years of experience in viral de novo sequencing projects.
- Facilitating high-quality genome reconstruction with the largest short and long-read sequencing capacity.
- Providing complete and customized solutions for in-depth analysis of viral genomes.
- Keeping up with the frontiers of microbial genomics and bioinformatics analysis.
Although most viral genomes do not require long reads to assemble, viruses exist as populations in infected hosts, and long-read sequencing technologies serve as powerful tools to adequately characterize these populations. For more information about our services, please fill out the inquiry form below. Our customer service representatives are available 24 hours a day from Monday to Sunday.
References
- Saud, Z., et al. (2021). "Nanopore sequencing and de novo assembly of a misidentified Camelpox vaccine reveals putative epigenetic modifications and alternate protein signal peptides." Scientific reports, 11(1), 1-13.
- Mizutani, Y., et al. (2021). "De novo Sequencing of Novel Mycoviruses From Fusarium sambucinum: An Attempt on Direct RNA Sequencing of Viral dsRNAs." Frontiers in microbiology, 12, 778.
- Castro, C. J., et al. (2020). "The effect of variant interference on de novo assembly for viral deep sequencing." BMC genomics, 21(1), 1-12.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment
