

We are dedicated to providing outstanding customer service and being reachable at all times.
Oxford Nanopore Sequencing Technology
As a leading genomics company, CD Genomics provides next-generation sequencing and bioinformatics services to pharmaceutical and biotech companies, as well as academic and government agencies around the world, relying on advanced sequencing instruments and rich project experience. The next era of DNA sequencing technology, also known as termed long-read sequencing or third-generation sequencing (TGS), plays an increasingly important role in biology. We are offering long-read sequencing services from PacBio and Oxford Nanopore, giving researchers a wide range of cutting-edge sequencing services to suit particular needs at every stage of any projects. With years of experience in long-read sequencing technology, we are able to generate and analyze high quality data using the latest workflows.
Oxford Nanopore Sequencing Technology (ONT)
-Technical Principle of ONT
Nanopore sequencing with Oxford Nanopore Technologies (ONT) systems is able to perform high-throughput long-read sequencing of both DNA and RNA samples, involving genomic DNA, amplified DNA, cDNA, and RNA. Briefly, the nanopore sequencer is based on a novel 'nanopore technology that uses nanopores embedded in an electro-resistant membrane. A voltage is set on the membrane to form an ionic current across the pores. When a DNA molecule tagged with tether proteinis guided and passes through a nanopore, the current variation, producing a characteristic signal, which is mainly influenced by four DNA bases. Then a base-calling algorithm converts the current variation back into the original DNA sequence in real-time.
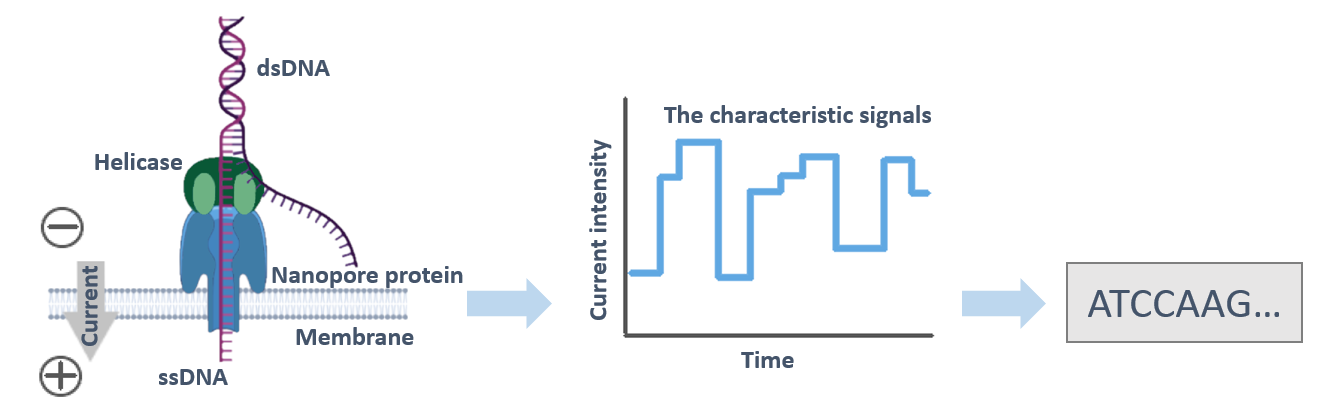 Fig.1 Schematic representation of a DNA molecule translocating a protein nanopore.
Fig.1 Schematic representation of a DNA molecule translocating a protein nanopore.
-Outstanding Advantages of ONT
- Long reads with no amplification
The long read length of over 150 Kbp provides a more comprehensive display of the sequence information of the target gene and facilitates the identification of duplicated gene fragments and structural variants. - Direct detection of target nucleic acids without PCR amplification
Enable direct sequencing of epigenetic modifications and RNA. - Real-time access to sequence information
Direct detection of the native strand of interest saves a lot of time. Sequencing signals are collected directly when the target nucleic acid passes through the nanopore, and sequencing results can be output in real time. - Simple library preparation
No PCR amplification, no fluorescent molecular markers, no optical imaging, extremely fast library preparation process.
-Applications of ONT
ONT directly sequences the native strand of interest, without optics or amplification and its data has been proven improve de novo genome assemblies, structural genomic variant, and so on. ONT has been applied in genomic analysis, identification of pathogens, metagenomic environmental monitoring, food safety monitoring and other aspects of life sciences.
Major ONT Systems are Available at CD Genomics

| Platforms | MinION | GridION | PromethION |
| Introductions | Each consumable flow cell can now generate 10 - 30Gb of DNA sequence data. | The GridION X5 is a compact benchtop system designed to run and analyze up to five MinION flow units. | PromethION offers the same real-time, long-read, direct DNA and RNA sequencing technology as MinION and GridION, but at a much larger scale. |
| Commonalities | -Real-time sequencing and analysis -Read length up to 2Mb |
||
| Differences | Pocket-Sized, portable device for biological analysis | High data output per run up to 5x20Gb | Super high data output per run up to 48x50Gb |
| Optimal Applications | -De novo sequencing -Targeted sequencing -Metagenomics -Epigenetics |
-De novo sequencing -Targeted sequencing -Metagenomics -Epigenetics |
-Population-scale sequencing -Plant genomics |
Based on Oxford Nanopore Sequencing technology, CD Genomics can help researchers and professionals with a wide range of cutting-edge sequencing services at competitive prices. Our clients have direct access to our staff and prompt feedback to their inquiries. If you are interested in our services, please don't hesitate to contact us for more details.
Related Services
- Human Genomics with Long-Read Sequencing
- Animal and Plant Genomics with Long-Read Sequencing
- Microbial Genomics with Long-Read Sequencing
- Transcriptomics with Long-Read Sequencing
- Epigenetics and Methylation Analysis Using Long-Read Sequencing
- Whole-Genome Resequencing with Long-Read Sequencing
- Pre-Made Library Long-Read Sequencing Services
- Long-Read Sequencing Data Analysis Services
References
- Noakes, M.T., et al, 2019. "Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage." Nature biotechnology, 37(6), pp.651-656.
- Kono, N. and Arakawa, K., 2019. "Nanopore sequencing: Review of potential applications in functional genomics." Development, growth & differentiation, 61(5), pp.316-326.
- Lin, B., Hui, J., & Mao, H. (2021). "Nanopore technology and its applications in gene sequencing." Biosensors, 11(7), 214.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment