

We are dedicated to providing outstanding customer service and being reachable at all times.
Oxford Nanopore Pre-Made Library Sequencing
The Oxford Nanopore Technologies (ONT) sequencing system uses a novel nanopore technology for sequencing long stretches of DNA or RNA. Unlike NGS methods that detect secondary signals, Oxford Nanopore sequencing technology directly identifies the current changes generated in real-time during the process. Changes in ionic currents are different for each nucleotide, producing a unique signature for each base. Then the electrical signal is translated into nucleotide bases (i.e. base-calling). ONT sequencing supports hundreds of kb of read length, greatly enhanced de novo genome assemblies, structural genomic variant, and transcriptome studies.
Nanopore Sequencing Services at CD Genomics

We are providing Nanopore sequencing services that are scalable from miniature devices to high-throughput installations. Different types of Nanopore sequencing are available, including,
-DNA sequencing, including long-read and ultra-long-read DNA sequencing.
-Full-length cDNA sequencing.
-Direct RNA sequencing.
Nanopore Sequencing Platforms
With the introduction of the Nanopore sequencing system, we have built a robust, scalable platform consisting of the MinION, GridION, and PromethION systems. These systems can now produce two major types of long reads with different lengths, accuracies, and throughputs, and can be used for specific applications.
Table 1. Data type, length, accuracy, and throughput across Oxford Nanopore sequencing platforms. (Logsdon, G. A., et al., 2020)
| Sequencing technology | Platform | Data type | Read length (kb) | Read accuracy (%) | Throughput per flow cell (Gb) | ||
| N50 | Maximum | Mean | Maximum | ||||
| Oxford Nanopore Technologies (ONT) | MinION/GridION | Long read | 10–60 | >1,000 | 87–98 | 2-20 | 30 |
| Ultra-long read | 100–200 | >1,500 | 0.5–2 | 2.5 | |||
| PromethION | Long read | 10–60 | >1,000 | 50–100 | 180 | ||
ONT Long and Ultra-Long Reads
Nanopore sequencing with ONT systems can yield huge read lengths due to its unique pore chemistry, which allows molecules to be transferred through the nanopore regardless of length. Here we outline the factors that affect the quality of ONT reads.
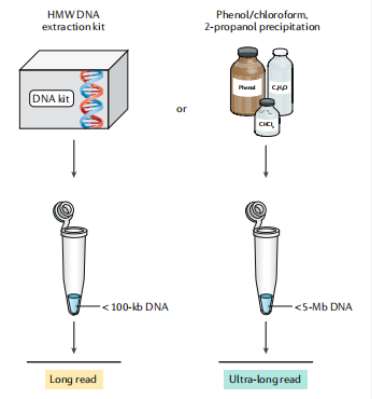 Fig2. ONT long and ultra-long reads. (Logsdon, G. A.,et al., 2020)
Fig2. ONT long and ultra-long reads. (Logsdon, G. A.,et al., 2020)
- ONT read lengths
The main factor limiting the length of ONT reads is considered to be the preparation of high molecular weight DNA, as has been demonstrated in various studies. Different DNA preparation and extraction methods are the basis of two main types of ONT data, namely standard long read (10-100kb) and specialized ultra-long read (>100kb). To generate standard long reads, nucleic acid extraction kits are often used, such as Qiagen's Puregene kit or Genomic-tip 500/G kit. While the traditional methods such as phenol-chloroform combined with ethanol or 2-propanol precipitation are recommended to extract HMW DNA for the generation of ultra-long reads. - ONT read accuracy
The consensus read accuracy of ONT long reads depends mainly on the use of the base-calling algorithm. Over the past few years, the accuracy of ONT reads has improved significantly due to these algorithm improvements. In order to obtain high accuracy ONT data (~97–98%), INC-seq, HiFRe, 1D2 sequencing, etc. are recommended.
Sample Requirements
- Genomic DNA samples, OD260/280=1.8~2.0, OD260/230=2.0~2.2, no degradation and no contamination
- Sample amount: Start with unamplified genomic DNA (>5µg input).
Thank you for your interest in our services. In order to get more details about our services, please contact us. We are glad to work with you!
References
- Logsdon, G. A., et al. (2020). "Long-read human genome sequencing and its applications." Nature Reviews Genetics, 21(10), 597-614.
- Mantere, T., et al. (2019). Long-read sequencing emerging in medical genetics. Frontiers in genetics, 10, 426.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment