

We are dedicated to providing outstanding customer service and being reachable at all times.
Nanopore RNA Sequencing

Long-read RNA-seq can overcome some of the inherent limitations of short-read RNA-seq to improve gene prediction and annotation in genome assembly, ultimately providing full-length RNA sequences and complete structural information. Besides Pacbio RNA sequencing services, CD Genomics also offers Oxford nanopore RNA sequencing services. Nanopore RNA sequencing technology enables accurate analysis of full-length native RNA or cDNA without the need to amplify or fragment, streamlining analysis and eliminating amplification bias and read length limitations. More importantly, it is currently the only technology capable of directly sequencing native RNA strands. Direct sequencing of RNA using nanopore sequencing technology identifies base modifications next to nucleotide sequences, analyzes poly-A tail lengths and transcription start sites, and analyzes full-length isoforms to recover the true RNA characteristics.
Features of Nanopore RNA Sequencing
- Generation of full-length transcripts, >20 kb single read transcripts.
- Direct RNA sequencing with amplification-free protocols and therefore no GC bias.
- Cost-effective identification of RNA base modifications.
- Provide transcript poly-A tail length estimations.
- Real-time sequencing and analysis for faster access to results.
- Platforms of different sizes to meet customer needs, such as Oxford MinION, GridION, or PromethION.
Oxford Nanopore Technology (ONT) Systems are All Available at CD Genomics
| Platforms | MinION | GridION | PromethION |
| Introductions | Each consumable flow cell can now generate 10 - 30Gb of DNA sequence data. | The GridION X5 is a compact benchtop system designed to run and analyze up to five MinION flow units. | PromethION offers the same real-time, long-read, direct DNA and RNA sequencing technology as MinION and GridION, but at a much larger scale. |
| Features | •Real-time sequencing and analysis •Pocket-Sized, portable device for biological analysis •Read length up to 2Mb |
•Real-time sequencing and analysis •Read length up to 2Mb •High data output per run-up to 5x20Gb |
•Real-time sequencing and analysis •Read length up to 2Mb •Super high data output per run-up to 48x50Gb |
Sequencing Strategies
- Sample type: Blood, tissues, cells, RNA.
- Library construction strategy and sample requirements:
-Direct RNA sequencing: RNA amount ≥1μg, OD260/280 =1.8 ~2.0.
-cDNA-PCR sequencing: RNA amount ≥ 500ng, OD260/280 =1.8 ~2.0.
-Direct cDNA sequencing: RNA amount ≥ 1μg, OD260/280 =1.8 ~2.0.
Workflow of Data Analysis
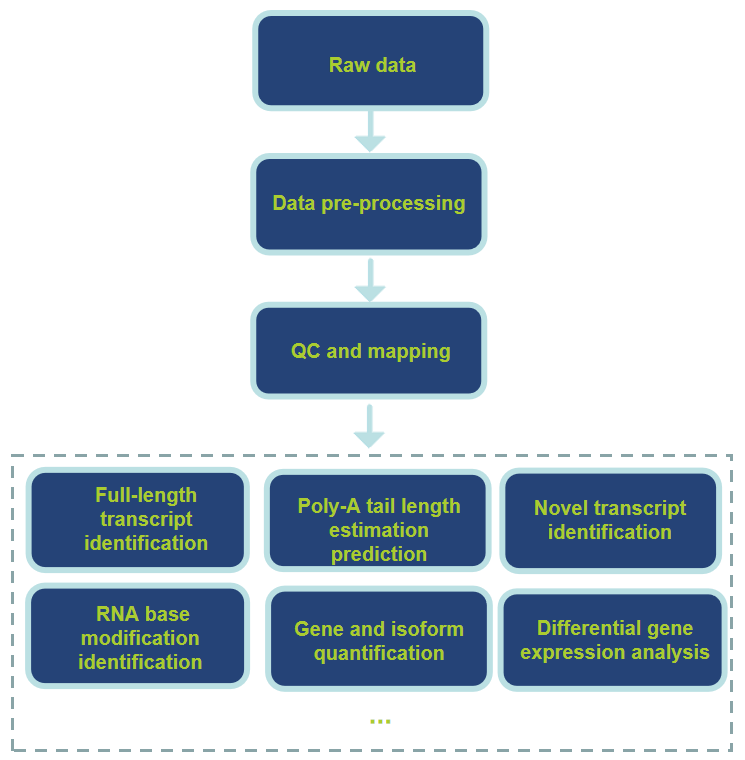
Benefits of Our Services
- No assembly is required. Long read lengths do not require assembly to obtain accurate sequence information of full-length transcripts.
- Comprehensive information analysis content. Keeping up with the frontier of scientific research, we constantly upgrade the information analysis content.
- Personalized analysis. With rich experience in personalized analysis, we can choose more suitable analysis software according to project needs to guarantee more accurate results.
- More new discoveries. New genes and transcriptional isoforms can be discovered and alternative splicing and gene fusion phenomena can be accurately identified.
Related Services
- Nanopore Direct RNA Sequencing
- Nanopore Full-Length cDNA Sequencing
- Animal/Plant RNA Sequencing
- Long-Read Sequencing of RNA Methylation
- Full-Length Transcriptome Profiling Combined with Long- and Short-Read Sequencing
- Full-Length Spatial-Temporal Transcriptome Profiling
- Single-Cell Full-Length Transcriptome Sequencing
Your questions and comments are important to us. Please feel free to contact us. We've got everything covered for your needs and looking forward to hearing from you.
References
- Cozzuto, L., et al. (2020). "MasterOfPores: a workflow for the analysis of oxford nanopore direct RNA sequencing datasets." Frontiers in genetics, 11, 211.
- Leonardi, T., & Leger, A. (2021). "Nanopore RNA sequencing analysis." In RNA Bioinformatics (pp. 569-578). Humana, New York, NY.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment