

We are dedicated to providing outstanding customer service and being reachable at all times.
Nanopore Full-Length cDNA Sequencing
Based on nanopore full-length cDNA sequencing, CD Genomics can help researchers to generate full-length transcript sequences containing the entire isomers, helping to more accurate identification of alternative splicing events and improve the analysis of transcriptional isoform expression.
Full-Length cDNA Sequencing
Full-length cDNA contains the complete sequence information of the respective mRNA templates. This information contains knowledge of the protein encoded by the original mRNA and the untranslated region (UTR) of that mRNA, which can be obtained by full-length cDNA sequencing.
As the most widely used high-throughput sequencing method today, short-read RNA sequencing (RNA-seq) has greatly expanded our ability to study transcriptome complexity. However, during sequencing, transcripts need to be cleaved into smaller fragments and then computationally reassembled due to short-read RNA-seq reads are only present a few hundred bases. As the number of alternative splice forms of genes increases, it becomes increasingly challenging to correctly assemble transcripts by computation. The disadvantages of short reads, such as uneven read coverage, complex splicing, and potential sequencing bias, will result in some information being lost from the original full-length transcripts.
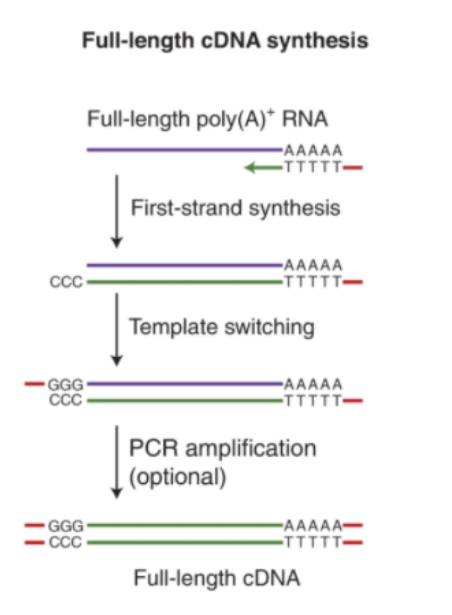 Fig1. Full-length cDNA synthesis for direct
Fig1. Full-length cDNA synthesis for direct
cDNA sequencing and PCR-cDNA sequencing.
(Wang, Y., et al., 2021)
Workflow of Our Full-Length cDNA Sequencing Service

Sample Requirements
-Sample type: Blood, tissues, cells, RNA samples (total RNA or poly-A+ RNA), and more.
-RNA amount ≥ 250ng, OD260/280 =1.8 ~2.0.
-RNA Integrity Number (RIN) ≥ 8.
Analysis Pipeline and Contents
-ONT sequencing data QC.
-Full-length transcript identification.
-Reference transcriptome alignment.
-Gene function and transcription structural annotation.
-Differential gene/transcription isoform quantification.
-Functional enrichment and protein interaction analysis.
Our Services Support
- Easy access to full-length transcript sequences, significantly reducing the rate of multi-locus comparisons.
- Cost-effective and accurate quantification of transcript levels.
- Accurate identification of splice variants and gene fusions.
- Real-time sequencing and analysis for faster access to results.
- Platforms of different sizes to meet customer needs, including MinION, GridION, or PromethION.
Correct expression of the full complexity of transcript isoforms will help in the discovery of important isoforms of disease states as well as help explore important agronomic traits. To perform full-length cDNA sequencing, researchers choose CD Genomics. With multiple specialists and years of experience in this field, we guarantee you high-quality data and integrated bioinformatics analyses. If you are not interested in our nanopore full-length cDNA sequencing, please feel free to contact us. We are still very happy to help you!
References
- Wang, Y., et al. (2021). "Nanopore sequencing technology, bioinformatics and applications." Nature biotechnology, 39(11), 1348-1365.
- Cui, J., et al. (2020). "Analysis and comprehensive comparison of PacBio and nanopore-based RNA sequencing of the Arabidopsis transcriptome." Plant Methods, 16(1), 1-13.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment