

We are dedicated to providing outstanding customer service and being reachable at all times.
Long-Read Sequencing of RNA Methylation
Emerging third-generation sequencing technologies (PacBio SMRT and Oxford Nanopore sequencing) can detect DNA and RNA methylation modifications directly. These newer sequencing technologies provide promising tools for further discovery of the mechanisms and functions of RNA methylation modifications. CD Genomics is based on the Oxford Nanopore sequencing platform to provide low-cost, high-efficiency, and high-accuracy RNA methylation identification and analysis. Our service enables direct identification of natural or synthetic RNA base modifications at nucleotide resolution, including N6-methyladenosine modification (m6A), 5-methylcytosine modification (m5C), and so forth.
The Importance of RNA Methylation
In recent years, with the development of high-throughput sequencing technology and the discovery of RNA methylation-related proteins, great progress has been made in RNA modification research. Nowadays, there are more than 150 RNA modifications have been detected and identified, and they have been found in messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and other noncoding small RNA (sncRNA).
RNA methylation is a kind of post-transcriptional regulation method. The major mammalian RNA methylation modifications include m6A, m5C, N1-methyladenosine (m1A), and pseudouridine modification (Ψ), but previous studies have mainly focused on m6A. Here, we give a brief overview of RNA m6A and its posttranscriptional regulation. m6A is the most common methylation modification in RNA, mainly distributed in long exons, stop codons, and in 3’UTRs. It is present in a variety of RNAs, with the highest abundance in mRNA. Similar to DNA methylation, m6A RNA methylation is characterized by dynamic and reversible in mammalian cells. As the most studied RNA methylation, m6A has been demonstrated that play an important role in RNA metabolism in terms of mRNA alternative polyadenylation, stability, and transport. Furthermore, m6A is closely associated with physiological processes such as cell differentiation, inflammation, and immunity. Aberrant m6A methylation has been reported to be implicated in obesity, cancer, neurodegeneration, and other pathologies.
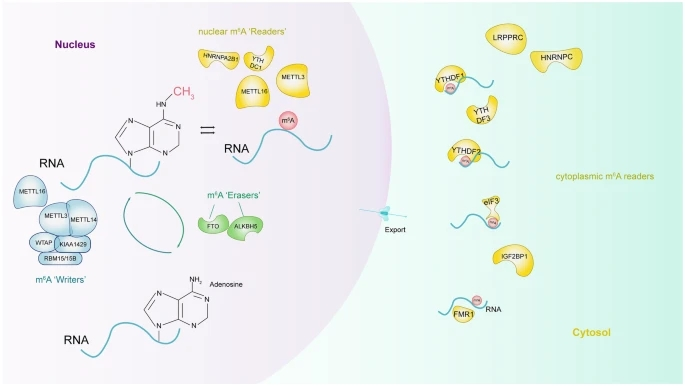 Fig1. Molecular composition of m6A RNA methylation. (Chen, X. Y., et al., 2019)
Fig1. Molecular composition of m6A RNA methylation. (Chen, X. Y., et al., 2019)
Benefits of RNA Methylation Profiling Using Long-Read Nanopore Sequencing
- No additional sample preparation is required.
- High efficiency and single-base resolution.
- A combination of high throughput and long-read length.
- Allows direct access to RNA methylation information.
- Greater experimental reproducibility.
Sample Requirements
-Sample type: conventional plant and animal samples.
-Recommended library inputs of 10 µg of RNA.
-Sequencing strategy: non-PCR amplification library construction, Nanopore PromethION platform.
Workflow of Our Service

Analysis Contents
- Data preprocessing and QC.
- Mapping reads to the reference genome.
- RNA modification site identification.
- Differential methylation analysis.
- Motif analysis.
- Functional analysis of RNA methylated genes.
CD Genomics is committed to providing an effective solution for assessing RNA methylation for animals and plants. If you are interested in our services, please don't hesitate to contact us. We've got everything covered for your needs and are ready to assist.
References
- Xu, L., & Seki, M. (2020). "Recent advances in the detection of base modifications using the Nanopore sequencer." Journal of human genetics, 65(1), 25-33.
- Parker, M. T., et al. (2020). "Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification." Elife, 9, e49658.
- Chen, X. Y., et al. (2019). "The role of m6A RNA methylation in human cancer." Molecular cancer, 18(1), 1-9.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment