

We are dedicated to providing outstanding customer service and being reachable at all times.
Long-Read Sequencing Data Analysis Services
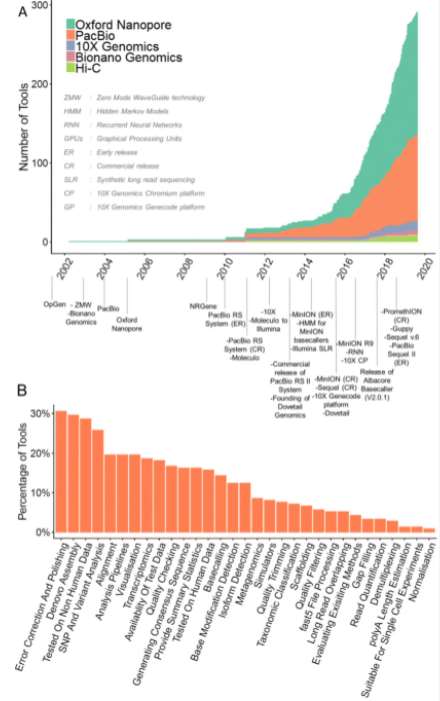 Fig1. overview of long-read analysis tools. (Amarasinghe, S. L., et al., 2020)
Fig1. overview of long-read analysis tools. (Amarasinghe, S. L., et al., 2020)
The use of long-read sequencing technologies has risen dramatically as sequencing costs have decreased and accuracy and throughput have continued to advance. CD Genomics is a leading global life sciences company. We specialize in the application of long-read sequencing technologies, including Oxford Nanopore and PacBio SMRT sequencing. Here, we are providing long-read sequencing data analysis in a wide range of fields such as whole-genome sequencing, RNA sequencing, epigenetics, and so on. Since long-read sequencing data analysis is a rapidly developing and advancing research area. Our data analysis uses the latest algorithms to provide a tailored solution to your specific application needs.
Overview of Long-Read Sequencing Analysis
Typically, long-read sequencing data analysis is a complex, computationally intensive, multi-step process that requires a high level of bioinformatics expertise. Each step in the analysis often requires a different set of tools or software. And some tools need significant computational resources, further adding to the complexity of the analysis process. As of early 2020, there are approximately 354 analytic tools for long-read data presented in publications (including preprints), social media, and online repositories. Among them, 262 tools are developed for performing Nanopore read analysis and 170 are SMRT data analysis tools. According to their functions, these tools can be divided into 31 groups, among which the more concerned and top-ranked are error correction polishing, de novo assembly, SNP and variant analysis.
Long-Read Sequencing Analysis Platform at CD Genomics
Main Applications of Our Long-Read Sequencing Data Analysis
We are committed to offering a flexible data analysis platform with diverse applications, including but not limited to the following:
- High-quality assembly of genomes.
- Isoform detection, differential gene expression and transcript.
- Structural variant and small variant calling analysis.
- Metagenomic classification and targeted 16S analysis.
- Modified base detection.
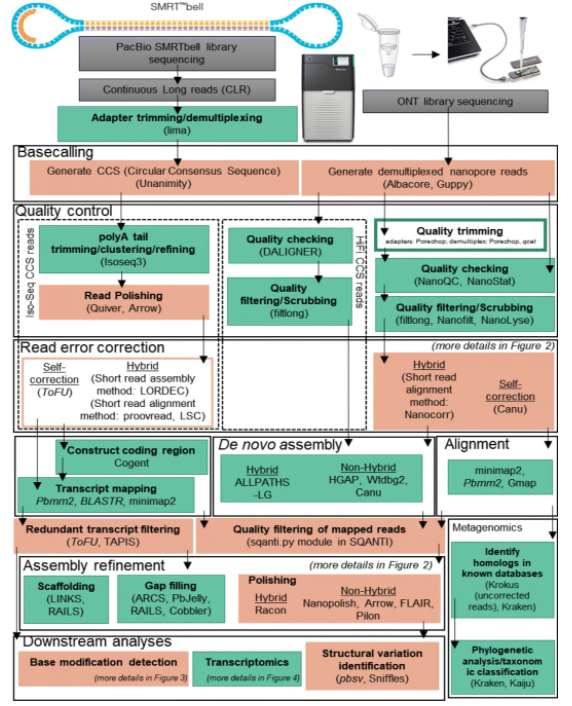 Fig 2. Typical long-read analysis pipelines for SMRT and nanopore data. (Amarasinghe, S. L., et al., 2020)
Fig 2. Typical long-read analysis pipelines for SMRT and nanopore data. (Amarasinghe, S. L., et al., 2020)
Benefits of Our Service
- Multiple analysis tools for different types of long-read sequencing data.
- Robust and customized analytical workflows.
- Dedicated bioinformaticians with extensive experience in long-read sequence data analysis.
- Provide intuitive, easy-to-use, and complete analytical study reports.
- Fast turnaround time with the most competitive prices.
Our long-read sequencing analysis platforms have integrated a series of long-read sequence tools. Please feel free to contact us for more information. We are glad to cooperate with you!
References
- Athanasopoulou, K., et al. (2021). "Third-Generation Sequencing: The Spearhead towards the Radical Transformation of Modern Genomics." Life, 12(1), 30.
- Amarasinghe, S. L., et al. (2020). Opportunities and challenges in long-read sequencing data analysis. Genome biology, 21(1), 1-16.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment