

We are dedicated to providing outstanding customer service and being reachable at all times.
Human Genome Structural Variation Detection
In order to gain a better view of structural variations (SVs), scientists and researchers have been trying to construct a catalog of human genome SVs using short-read sequencing and long-read technologies. However, the limited length of short-read technologies hinders the detection of SVs. The short reads usually lack sensitivity, especially for insertion detection. Long-read sequencing technologies have improved throughput and accuracy and reduced cost per Mb, providing the best option for structural variance detection in the genome. CD Genomics is a leading global life sciences company. Based on the third-generation sequencing platforms, Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), we are capable of helping our customers comprehensively detect SVs at a higher resolution.
Overview of Structural Variations (SVs)
Over the past two decades, scientists have realized the importance of SVs for genetic variation among individuals and that their effect is greater at the nucleotide level than single nucleotide polymorphisms (SNPs) and short insertion-deletion mutations (InDels) together. SVs represent genomic rearrangements, including large insertions, deletions, inversions, duplications, translocations, and complex combinations of these mutations, with a length of at least 50 bp. As the largest form of genetic variation in the human genome, SVs are closely associated with gene regulations, evolution, phenotype, and human diseases ( e.g. cancers, neurological disorders and inherited diseases).
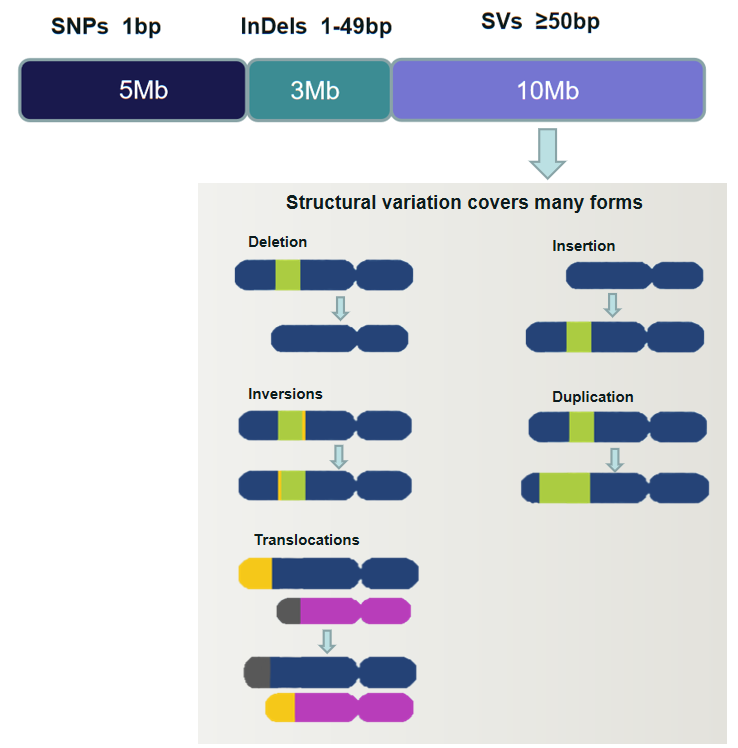
Advantages of Using Long-Read Sequencing Technology for Detecting SVs
Long-read sequencing technology, also known as third-generation sequencing technology, with the advantages of long-read length, no PCR process, can easily span genomic repetitive regions, high GC regions, etc., to make up for the missed detection of SVs by traditional short-read sequencing technology, providing an advantage potential to increase the reliability and resolution of SV detection in the human genome.
- Due to the length of longreads (> 10 kb), long reads can be calibrated with high confidence between SV breakpoints.
- Long read can be used to identify complex SVs that were missed when using short-read data.
We provide a complete end-to-end solution of our projects, including

Sample Requirements
- Start with unamplified genomic DNA (>5µg input).
- Other sample types, including blood, tissue, cell lines, and so on.
Long-read SV Calling Analysis Workflow
- PacBio or ONT read quality control (QC).
- Align reads to hg38 (Minimap2, BLASR, and NGMLR).
- SV detection (Sniffles, SMRT-SV, PB-Honey, and NanoSV) and sorting (VCFsort).
- Create a summary of the variant calls (awk).
Advantage of Our Services
- PacBio and ONT library preparation and barcoding.
- The Sequel II sequencer and the PromethION device (achieve >15-fold coverage of a human genome).
- SV analysis using an optimized pipeline with state-of-the-art tools.
- Identification of the effects of SVs. In addition to SV detection, the epigenome status (including haplotype and DNA methylation information) surrounding SV loci can also be investigated.
- Lowest price (note that prices depend on sample size and quantity, please contact us for a quotation).
In addition, we can customize our services to meet your project needs, including DNA isolation, raw data transfer, visualization tools to deliver results, increased coverage, and more. In contrast to short-read sequencing, long-read sequencing has lower false-positive and false-negative rates, making it preferred to accurately detect and fully resolve SVs. For more information on how we can help you, please feel free to contact us.
References
- Sakamoto, Y., et al. (2021). "Application of long-read sequencing to the detection of structural variants in human cancer genomes." Computational and Structural Biotechnology Journal, 19, 4207-4216.
- Jiang, T., et al. (2020). "Long-read-based human genomic structural variation detection with cuteSV." Genome biology, 21(1), 1-24.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment