

We are dedicated to providing outstanding customer service and being reachable at all times.
Genome-Wide Association Study (GWAS)
By combining the phenotypic data of the target trait and performing genome-wide association analysis based on certain statistical methods, the chromosomal segments or loci affecting the phenotypic variation of the target trait can be quickly obtained. Based on long-read sequencing platforms at CD Genomics, researchers and professionals can quickly, accurately, and efficiently localize genes for multiple target traits.
Application of Genome-Wide Association Study (GWAS)

Genome-wide association study (GWAS) is a method to analyze the association between molecular marker data and phenotypic traits using a high density of genome-wide genetic markers for the population under study. In the past two decades, GWAS has become an effective tool for studying the genetic structure of complex traits in humans, animals, and plants. More than 6,000 variant loci associated with > 500 quantitative traits and complex diseases have been identified in humans. In animals, GWAS has been applied to complex diseases such as economically important traits and genetic defective diseases in major livestock and poultry. On the plant side, GWAS analysis has been performed for flowering time, developmental traits, and agronomic traits in Arabidopsis, rice, maize, and cotton.
Service Offering at CD Genomics
SV-GWAS is the use of long-read resequencing techniques to obtain information on structural variations (SVs) in a population and screening for SVs associated with the target traits. Here, we combined long-read sequencing with the results of the next-generation sequencing-based GWAS analysis to achieve more accurate trait-associated gene localization.
Sample Requirements
- Sample Quantity: ≥200 samples (WGS); select at least 10 representative individuals for long-read sequencing.
- DNA amount:
-Illumina platform: ≥ 1 μg. OD260/280=1.8~2.0, OD260/230=2.0~2.2, no degradation and no contamination
-PacBio/ONT platform: ≥ 10 μg. OD260/280=1.8~2.0, OD260/230=2.0~2.2, no degradation and no contamination - Sequencing strategy:
-Illumina platform: 350 bp library, ≥ 5 X.
-PacBio/ONT platform: ≥10kb, ≥ 30X.
Workflow of Our Service

Analysis Workflow
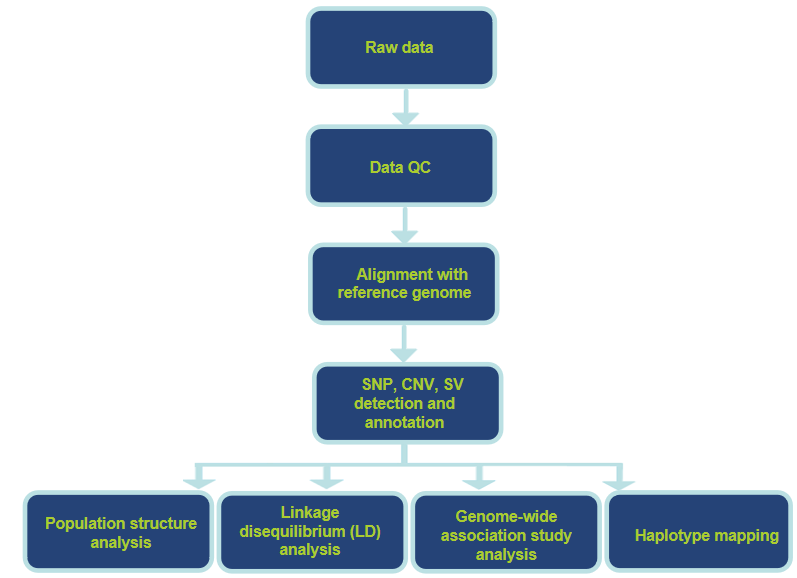
Specific Analysis Contents
- Data quality control.
- SNP, CNV, SV detection and annotation.
- Genome-wide SNP enrichment analysis.
- Differential SV analysis.
- Population SV typing.
- Phylogenetic tree construction.
- Principal component analysis (PAC).
- Linkage disequilibrium (LD) analysis.
- Marker-trait association analysis.
- Functional annotation of genes in target trait-related regions.
- Haplotype mapping.
Benefits of Our Service
- Genome-wide level, more complete, and denser variation information.
- Enhanced SV-GWAS analysis with long-read.
- High resolution up to the single-base level.
- Precise localization of candidate genes or gene regions associated with traits.
GWAS refers to a statistical approach in genetics to identify variants that correlate with a certain phenotype. GWAS opens a new chapter in genomics and genetics research, linking genomics and genetics on an unprecedented scale. CD Genomics is a leading global life sciences company. We specialize in the application of third-generation sequencing technologies. If you are interested in our GWAS with long-read sequencing, please feel free to contact us. We are looking forward to cooperating with you.
References
- Mantere, T., et al. (2019). "Long-read sequencing emerging in medical genetics." Frontiers in genetics, 10, 426.
- Cano-Gamez, E., & Trynka, G. (2020). "From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases." Frontiers in genetics, 11, 424.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment