

We are dedicated to providing outstanding customer service and being reachable at all times.
Fungal Whole-Genome De Novo Sequencing
In addition to supporting the de novo assembly of bacterial genomes, CD Genomics also offers fungal whole-genome de novo sequencing services based on advanced long-read sequencing technologies. Our services enable the advanced assembly of fungi and provide powerful support for fungal research, which can be used to predict important genes and proteins of fungi to understand their functions and possible mechanisms.
De Novo Assembly of Fungal Genomes
Fungi are diverse and differ in morphology, physiology, and pathogenicity. In order to study and reveal the origins of the diversities in these traits, molecular genetic studies are needed. Identification of species and pathogenicity levels of fungi is crucial for fungi research, such as resolving complex phylogeny of fungi. The genomic data can facilitate the understanding of their impact on the environment and the host-pathogen interactions. In addition, fungi are closely related to human life. Fungal genomic data is also important for industrial applications. With advances in sequencing technologies, more accurate, longer, and more affordable long reads are available. De novo genome assembly based on long reads continues to evolve rapidly and has been a powerful method to assemble complex microbial genomes, including fungal genomes.
Fungal Whole Genome De Novo Sequencing Service Overview
| Sequencing Platform | Illumina HiSeq PE150 | PacBio Sequel ll | Nanopore PromethION |
| Sample Requirements | Sample Type: genomic DNA Amount: ≥ 200ng OD260/280=1.8~2.0 no degradation and no contamination |
Sample Type: HMW genomic DNA Amount: ≥ 15μg OD260/280=1.7~2.0 Fragments should be ≥ 20 kb |
Sample Type: HMW genomic DNA Amount: ≥ 10μg OD260/280=1.75~2.0 Fragments should be ≥ 20 kb |
| Sequencing Strategies | 350 bp insert library, ≥ 50 X | 20Kb SMRTbell library, CLR sequencing, ≥100X | ONT Library preparation, ≥100X |
| Application Scope | Large-scale sequencing and rapid scanning | Sequencing of a small number of strains Accuracy in place |
Sequencing of a small number of strains Accuracy in place |
| Delivery Cycle | Minimum of 30 working days | Minimum of 45 working days | Minimum of 40 working days |
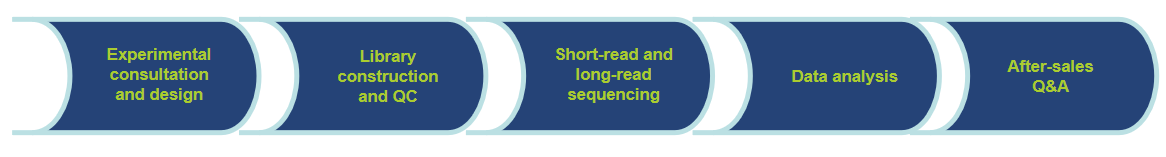
Workflow of Our Services
Analysis Contents
Analysis of fungal genome information at different levels of fineness, deciphering the development of fungi and exploring their potential value. Our analysis consists of two main parts: standard analysis and advanced analysis. Specific analytical services are as follows, but are not limited to,
- Standard analysis
-Data quality control.
-Genome assembly and evaluation of assembly results.
-Genome component analysis.
-Gene functional annotation. - Advanced analysis
-Gene family construction.
-Phylogenetic analysis.
-Gene synteny analysis.
-Pathogen-host interaction (PHI) database annotation.
-CAZy database annotation.
-Fungal virulence factor (DFVF) analysis.
-Identification of novel secondary metabolite biosynthesis (SMB) genes.
Fungal species are large and diverse, with relatively simple genomes. The complete assembly and accurate annotation of fungal genomes will provide the basis for fungal diversity and evolution studies, host-fungal interaction mechanisms, and antifungal drug development. CD Genomics provides end-to-end and customized services. Based on the advanced platform and years of experience in microbial de novo sequencing services, we provide you with high-quality data, and reliable and cutting-edge bioinformatics analysis. For more information about our services, please just complete the inquiry form below.
References
- McKenzie, S. K., et al. (2020). "Complete, high-quality genomes from long-read metagenomic sequencing of two wolf lichen thalli reveals enigmatic genome architecture." Genomics, 112(5), 3150-3156.
- Krasnov, G. S., et al. (2020). "High-quality genome assembly of Fusarium oxysporum f. sp. lini." Frontiers in genetics, 959.
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment
