

We are dedicated to providing outstanding customer service and being reachable at all times.
Full-Length Plasmid Sequencing
Bacterial plasmids have been widely sequenced and serve as an essential tool in modern molecular biology research. CD Genomics is proud to offer full-length plasmid sequencing services based on two sequencing strategies, including the PacBio SMRT sequencing platform, and a combined Illumina sequencing and Oxford Nanopore Technologies (ONT) Nanopore sequencing strategy. Our services are available to meet the specific needs of our clients. Based on accuracy and affordability, our experts will provide them with the most suitable strategy to achieve their research goals.
Overview of Full-Length Plasmid Sequencing
Plasmids are genetic materials that have the ability to replicate independently outside the bacterial chromosome. Most plasmids are loop-closed, but linear dsDNA exists, ranging in size from 5 to 500kb. Although smaller than chromosomes, plasmids are important genomic components, providing the bacterial host with additional genetic material that is important for its survival and adaptation. Plasmids carry genetic information that can confer certain specific phenotypic traits such as fertility, drug resistance, pathogenicity, and metabolic alterations to the host bacteria and facilitate intra/interspecies gene flow.
Next-generation sequencing (NGS) technologies (short-read sequencing) have been widely applied to perform whole-genome analysis. However, assembling accessory regions of the genome using short-read NGS data is a challenge. Long sequence sequencing, including PacBio SMRT sequencing and ONT nanopore sequencing, can generate long sequence sequences, which have great advantages for genome assembly, especially plasmid assembly.
Our Full-Length Plasmid Sequencing Service
- PacBio SMRT sequencing
PacBio SMRT sequencing represents the first long-read sequencing that generates reads with an average length of 11 kb. The method has high sequence coverage (>100×) and high accuracy on single nucleotides but is relatively costly. - A strategy combining Illumina sequencing and ONT Nanopore sequencing
This method combines long-read assembly and short-read error correction. The accuracy of nanopore sequencing is generally reported to be 90%, and some researchers have reported that sequencing accuracy can reach 99.96% after read polishing. - Service details
-DNA amount: ≥200 ng
-Highly sensitive detection of contaminating DNA sequences
-No reference sequence is required
-Turnaround time depends on the size and number of samples.
Workflow of Our Service

Technology Pipeline
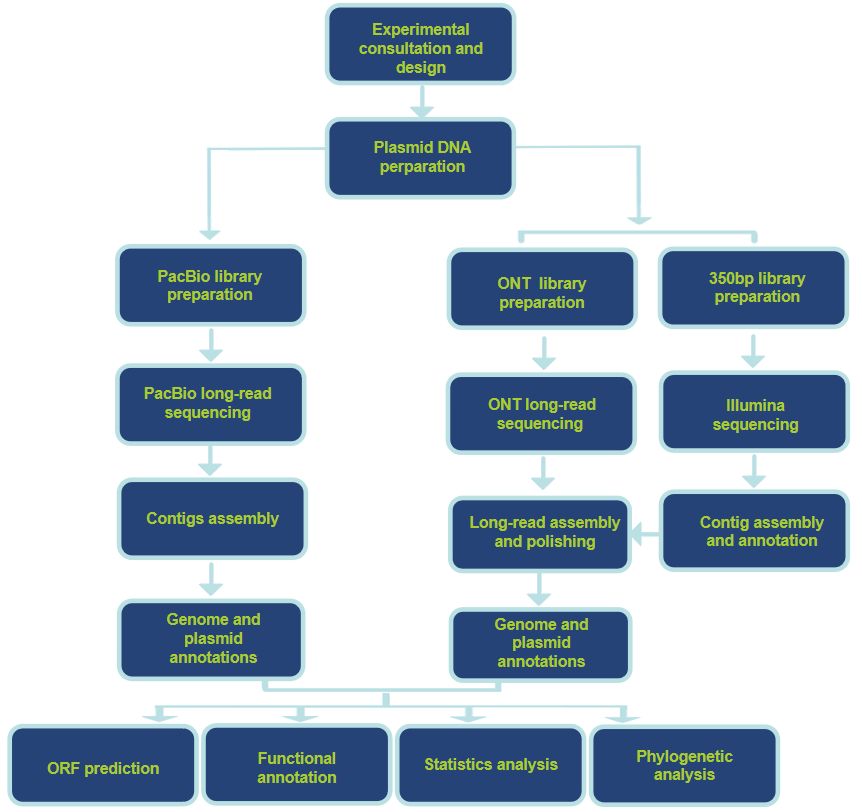
Analysis Contents
- Data re-processing and QC
- Contig assembly and annotation
- Genome and plasmid annotations
- Detection and identification of virulence genes
- Detection and identification of antimicrobial genes
- Phylogenetic analysis
- Comparative analysis
To obtain complete and accurate plasmid information, researchers and professionals choose CD Genomics. Our service workflow is robust and powerful and is capable of recovering small plasmid sequences from complex microbiomes. We promise that each plasmid is fully sequenced by long reads with a high level of consistent accuracy. Please don't hesitate to contact us. We've got everything covered for your needs and are ready to assist.
References
- Taylor, T. L., et al. (2019). "Rapid, multiplexed, whole genome and plasmid sequencing of foodborne pathogens using long-read nanopore technology." Scientific reports, 9(1), 1-11.
- Wick, R. R., et al. (2021). "Recovery of small plasmid sequences via Oxford Nanopore sequencing." Microbial genomics, 7(8).
For research purposes only, not intended for personal diagnosis, clinical testing, or health assessment
